Loading Task Data
Contents
Loading Task Data#
(version with Behavioural Data)#
The HCP dataset comprises task-based fMRI from a large sample of human subjects. The NMA-curated dataset includes time series data that has been preprocessed and spatially-downsampled by aggregating within 360 regions of interest.
In order to use this dataset, please electronically sign the HCP data use terms at ConnectomeDB. Instructions for this are on pp. 24-25 of the HCP Reference Manual.
In this notebook, NMA provides code for downloading the data and doing some basic visualisation and processing.
# @title Install dependencies
!pip install nilearn --quiet
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 10.3/10.3 MB 15.7 MB/s eta 0:00:00
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 3.3/3.3 MB 19.7 MB/s eta 0:00:00
?25h
import os
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
#@title Figure settings
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
plt.style.use("https://raw.githubusercontent.com/NeuromatchAcademy/course-content/main/nma.mplstyle")
# The data shared for NMA projects is a subset of the full HCP dataset
N_SUBJECTS = 100
# The data have already been aggregated into ROIs from the Glasser parcellation
N_PARCELS = 360
# The acquisition parameters for all tasks were identical
TR = 0.72 # Time resolution, in seconds = Sampling rate
# The parcels are matched across hemispheres with the same order
HEMIS = ["Right", "Left"]
# Each experiment was repeated twice in each subject
RUNS = ['LR','RL']
N_RUNS = 2
# There are 7 tasks. Each has a number of 'conditions'
# TIP: look inside the data folders for more fine-graned conditions
EXPERIMENTS = {
'MOTOR' : {'cond':['lf','rf','lh','rh','t','cue']},
'WM' : {'cond':['0bk_body','0bk_faces','0bk_places','0bk_tools','2bk_body','2bk_faces','2bk_places','2bk_tools']},
'EMOTION' : {'cond':['fear','neut']},
'GAMBLING' : {'cond':['loss','win']},
'LANGUAGE' : {'cond':['math','story']},
'RELATIONAL' : {'cond':['match','relation']},
'SOCIAL' : {'cond':['mental','rnd']}
}
For a detailed description of the tasks have a look pages 45-54 of the HCP reference manual.
Downloading data#
The task data are shared in different files, but they will unpack into the same directory structure.
# @title Download data file
import os, requests
fname = "hcp_task.tgz"
url = "https://osf.io/2y3fw/download"
# urls = ["https://osf. io/bqp7m/download"
# "https://osf. io/s4h8j/download"
# "https://osf. io/x5p4g/download"
# "https://osf. io/j5kuc/download"
# "https://osf.io/s4h8i/download/" Bati provided this
if not os.path.isfile(fname):
try:
r = requests.get(url)
except requests.ConnectionError:
print("!!! Failed to download data !!!")
else:
if r.status_code != requests.codes.ok:
print("!!! Failed to download data !!!")
else:
with open(fname, "wb") as fid:
fid.write(r.content)
# The download cells will store the data in nested directories starting here:
HCP_DIR = "./hcp_task"
# importing the "tarfile" module
import tarfile
# open file
with tarfile.open(fname) as tfile:
# extracting file
tfile.extractall('.')
subjects = np.loadtxt(os.path.join(HCP_DIR, 'subjects_list.txt'), dtype='str')
Understanding the folder organisation#
The data folder has the following organisation:
hcp
regions.npy (information on the brain parcellation)
subjects_list.txt (list of subject IDs)
subjects (main data folder)
[subjectID] (subject-specific subfolder)
EXPERIMENT (one folder per experiment)
RUN (one folder per run)
data.npy (the parcellated time series data)
EVs (EVs folder)
[ev1.txt] (one file per condition)
[ev2.txt]
Stats.txt (behavioural data [where available] - averaged per run)
Sync.txt (ignore this file)
Loading region information#
Downloading this dataset will create the regions.npy file, which contains the region name and network assignment for each parcel.
Detailed information about the name used for each region is provided in the Supplement to Glasser et al. 2016.
Information about the network parcellation is provided in Ji et al, 2019.
regions = np.load(f"{HCP_DIR}/regions.npy").T
region_info = dict(
name=regions[0].tolist(),
network=regions[1],
hemi=['Right']*int(N_PARCELS/2) + ['Left']*int(N_PARCELS/2),
)
#list of unique network name
unique_net = []
for net in region_info['network']:
if net not in unique_net:
unique_net.append(net)
print("there are ",len(unique_net),"unique networks.")
print(unique_net)
there are 12 unique networks.
['Visual1', 'Visual2', 'Somatomotor', 'Cingulo-Oper', 'Language', 'Default', 'Frontopariet', 'Auditory', 'Dorsal-atten', 'Posterior-Mu', 'Orbito-Affec', 'Ventral-Mult']
#list of regions in each network
dict_reg_in_net = dict()
for net in unique_net:
dict_reg_in_net[net] = [] #establish empty list associate with key 'str of network'
for idx in range(len(region_info['name'])):
current_brain_reg = region_info['name'][idx]
if region_info['network'][idx] == net and (current_brain_reg not in dict_reg_in_net[net]):
dict_reg_in_net[net].append(current_brain_reg)
for network,list_reg in dict_reg_in_net.items():
print("Network name: ", network, "contains ", len(list_reg)," regions:")
print(list_reg)
Network name: Visual1 contains 6 regions:
['R_V1', 'R_ProS', 'R_DVT', 'L_V1', 'L_ProS', 'L_DVT']
Network name: Visual2 contains 54 regions:
['R_MST', 'R_V6', 'R_V2', 'R_V3', 'R_V4', 'R_V8', 'R_V3A', 'R_V7', 'R_IPS1', 'R_FFC', 'R_V3B', 'R_LO1', 'R_LO2', 'R_PIT', 'R_MT', 'R_LIPv', 'R_VIP', 'R_PH', 'R_V6A', 'R_VMV1', 'R_VMV3', 'R_V4t', 'R_FST', 'R_V3CD', 'R_LO3', 'R_VMV2', 'R_VVC', 'L_MST', 'L_V6', 'L_V2', 'L_V3', 'L_V4', 'L_V8', 'L_V3A', 'L_V7', 'L_IPS1', 'L_FFC', 'L_V3B', 'L_LO1', 'L_LO2', 'L_PIT', 'L_MT', 'L_LIPv', 'L_VIP', 'L_PH', 'L_V6A', 'L_VMV1', 'L_VMV3', 'L_V4t', 'L_FST', 'L_V3CD', 'L_LO3', 'L_VMV2', 'L_VVC']
Network name: Somatomotor contains 39 regions:
['R_4', 'R_3b', 'R_5m', 'R_5L', 'R_24dd', 'R_24dv', 'R_7AL', 'R_7PC', 'R_1', 'R_2', 'R_3a', 'R_6d', 'R_6mp', 'R_6v', 'R_OP4', 'R_OP1', 'R_OP2-3', 'R_FOP2', 'R_Ig', 'L_4', 'L_3b', 'L_5m', 'L_5L', 'L_24dd', 'L_24dv', 'L_7AL', 'L_7PC', 'L_1', 'L_2', 'L_3a', 'L_6d', 'L_6mp', 'L_6v', 'L_OP4', 'L_OP1', 'L_OP2-3', 'L_RI', 'L_FOP2', 'L_Ig']
Network name: Cingulo-Oper contains 56 regions:
['R_FEF', 'R_5mv', 'R_23c', 'R_SCEF', 'R_6ma', 'R_7Am', 'R_p24pr', 'R_33pr', 'R_a24pr', 'R_p32pr', 'R_6r', 'R_46', 'R_9-46d', 'R_43', 'R_PFcm', 'R_PoI2', 'R_FOP4', 'R_MI', 'R_FOP1', 'R_FOP3', 'R_PFop', 'R_PF', 'R_PoI1', 'R_FOP5', 'R_PI', 'R_a32pr', 'R_p24', 'L_FEF', 'L_PEF', 'L_PSL', 'L_5mv', 'L_23c', 'L_SCEF', 'L_6ma', 'L_7Am', 'L_p24pr', 'L_a24pr', 'L_p32pr', 'L_6r', 'L_IFSa', 'L_46', 'L_9-46d', 'L_43', 'L_PFcm', 'L_PoI2', 'L_FOP4', 'L_MI', 'L_FOP1', 'L_FOP3', 'L_PFop', 'L_PF', 'L_PoI1', 'L_FOP5', 'L_PI', 'L_a32pr', 'L_p24']
Network name: Language contains 23 regions:
['R_PEF', 'R_7PL', 'R_MIP', 'R_LIPd', 'R_6a', 'R_PFt', 'R_AIP', 'R_PHA3', 'R_TE2p', 'R_PHT', 'R_PGp', 'R_IP0', 'L_7PL', 'L_MIP', 'L_LIPd', 'L_6a', 'L_PFt', 'L_AIP', 'L_PHA3', 'L_TE2p', 'L_PHT', 'L_PGp', 'L_IP0']
Network name: Default contains 23 regions:
['R_55b', 'R_PSL', 'R_SFL', 'R_STV', 'R_44', 'R_45', 'R_IFJa', 'R_IFSp', 'R_STGa', 'R_A5', 'R_STSda', 'R_STSdp', 'R_TPOJ1', 'R_TGv', 'L_55b', 'L_SFL', 'L_45', 'L_IFJa', 'L_STGa', 'L_A5', 'L_STSdp', 'L_TPOJ1', 'L_TGv']
Network name: Frontopariet contains 50 regions:
['R_RSC', 'R_POS2', 'R_7Pm', 'R_8BM', 'R_8C', 'R_a47r', 'R_IFJp', 'R_IFSa', 'R_p9-46v', 'R_a9-46v', 'R_a10p', 'R_11l', 'R_13l', 'R_i6-8', 'R_s6-8', 'R_AVI', 'R_TE1p', 'R_IP2', 'R_IP1', 'R_PFm', 'R_p10p', 'R_p47r', 'L_RSC', 'L_POS2', 'L_7Pm', 'L_33pr', 'L_d32', 'L_8BM', 'L_8C', 'L_44', 'L_a47r', 'L_IFJp', 'L_IFSp', 'L_p9-46v', 'L_a9-46v', 'L_a10p', 'L_11l', 'L_13l', 'L_OFC', 'L_i6-8', 'L_s6-8', 'L_AVI', 'L_TE1p', 'L_IP2', 'L_IP1', 'L_PFm', 'L_31a', 'L_p10p', 'L_p47r', 'L_TE1m']
Network name: Auditory contains 15 regions:
['R_A1', 'R_52', 'R_RI', 'R_TA2', 'R_PBelt', 'R_MBelt', 'R_LBelt', 'R_A4', 'L_A1', 'L_52', 'L_TA2', 'L_PBelt', 'L_MBelt', 'L_LBelt', 'L_A4']
Network name: Dorsal-atten contains 7 regions:
['R_PCV', 'R_TPOJ2', 'R_TPOJ3', 'L_PCV', 'L_STV', 'L_TPOJ2', 'L_TPOJ3']
Network name: Posterior-Mu contains 77 regions:
['R_7m', 'R_POS1', 'R_23d', 'R_v23ab', 'R_d23ab', 'R_31pv', 'R_a24', 'R_d32', 'R_p32', 'R_10r', 'R_47m', 'R_8Av', 'R_8Ad', 'R_9m', 'R_8BL', 'R_9p', 'R_10d', 'R_47l', 'R_9a', 'R_10v', 'R_10pp', 'R_OFC', 'R_47s', 'R_EC', 'R_PreS', 'R_H', 'R_PHA1', 'R_STSvp', 'R_TGd', 'R_TE1a', 'R_TE2a', 'R_PGi', 'R_PGs', 'R_PHA2', 'R_31pd', 'R_31a', 'R_25', 'R_s32', 'R_STSva', 'R_TE1m', 'L_7m', 'L_POS1', 'L_23d', 'L_v23ab', 'L_d23ab', 'L_31pv', 'L_a24', 'L_p32', 'L_10r', 'L_47m', 'L_8Av', 'L_8Ad', 'L_9m', 'L_8BL', 'L_9p', 'L_10d', 'L_47l', 'L_9a', 'L_10v', 'L_10pp', 'L_47s', 'L_EC', 'L_PreS', 'L_H', 'L_PHA1', 'L_STSda', 'L_STSvp', 'L_TGd', 'L_TE1a', 'L_TE2a', 'L_PGi', 'L_PGs', 'L_PHA2', 'L_31pd', 'L_25', 'L_s32', 'L_STSva']
Network name: Orbito-Affec contains 6 regions:
['R_Pir', 'R_AAIC', 'R_pOFC', 'L_Pir', 'L_AAIC', 'L_pOFC']
Network name: Ventral-Mult contains 4 regions:
['R_PeEc', 'R_TF', 'L_PeEc', 'L_TF']
Examining the regions#
#examine the regions by myself
print(len(regions))
print(regions)
print(len(region_info['name']))
print(region_info.keys())
print(region_info['hemi'])
3
[['R_V1' 'R_MST' 'R_V6' ... 'L_PI' 'L_a32pr' 'L_p24']
['Visual1' 'Visual2' 'Visual2' ... 'Cingulo-Oper' 'Cingulo-Oper'
'Cingulo-Oper']
['2.209' '2.05561' '2.1498' ... '1.74335' '1.73082' '1.65968']]
360
dict_keys(['name', 'network', 'hemi'])
['Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Right', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left', 'Left']
Help functions#
We provide two helper functions: one for loading the time series from a single suject and a single run, and one for loading an EV file for each task.
An EV file (EV:Explanatory Variable) describes the task experiment in terms of stimulus onset, duration, and amplitude. These can be used to model the task time series data.
def load_single_timeseries(subject, experiment, run, remove_mean=True):
"""Load timeseries data for a single subject and single run.
Args:
subject (str): subject ID to load
experiment (str): Name of experiment
run (int): (0 or 1)
remove_mean (bool): If True, subtract the parcel-wise mean (typically the mean BOLD signal is not of interest)
Returns
ts (n_parcel x n_timepoint array): Array of BOLD data values
"""
bold_run = RUNS[run]
bold_path = f"{HCP_DIR}/subjects/{subject}/{experiment}/tfMRI_{experiment}_{bold_run}"
bold_file = "data.npy"
ts = np.load(f"{bold_path}/{bold_file}") #ts is timeseries
# print("this is ts",ts)
# print("len(t)",len(ts))
# print("ts.mean(axis=1, keepdims=True)", ts.mean(axis=1, keepdims=True))
if remove_mean:
ts -= ts.mean(axis=1, keepdims=True)
return ts
def load_evs(subject, experiment, run):
"""Load EVs (explanatory variables) data for one task experiment.
Args:
subject (str): subject ID to load
experiment (str) : Name of experiment
run (int): 0 or 1
Returns
evs (list of lists): A list of frames associated with each condition
"""
frames_list = []
task_key = f'tfMRI_{experiment}_{RUNS[run]}'
for cond in EXPERIMENTS[experiment]['cond']:
ev_file = f"{HCP_DIR}/subjects/{subject}/{experiment}/{task_key}/EVs/{cond}.txt"
ev_array = np.loadtxt(ev_file, ndmin=2, unpack=True)
# print("ev_array",ev_array) ; basically just array of what's in txt file
ev = dict(zip(["onset", "duration", "amplitude"], ev_array))
# Determine when trial starts, rounded down
start = np.floor(ev["onset"] / TR).astype(int) # this is start index for each frame
# Use trial duration to determine how many frames to include for trial
duration = np.ceil(ev["duration"] / TR).astype(int) # this is **how many** indices for that frame
# Take the range of frames that correspond to this specific trial
frames = [s + np.arange(0, d) for s, d in zip(start, duration)]
frames_list.append(frames)
return frames_list
Example run (EMOTION task)#
Let’s load the timeseries data for the MOTOR experiment from a single subject and a single run
my_exp = 'EMOTION'
my_subj = subjects[0]
my_run = 1
data = load_single_timeseries(subject=my_subj,
experiment=my_exp,
run=my_run,
remove_mean=True)
print(data.shape)
(360, 176)
As you can see the time series data contains 284 time points in 360 regions of interest (ROIs).
Now in order to understand how to model these data, we need to relate the time series to the experimental manipulation. This is described by the EV files. Let us load the EVs for this experiment.
evs = load_evs(subject=my_subj, experiment=my_exp, run=my_run)
Trying to understand evs (archived)#
print("frames of first cond:",evs[0])
print("frames of second cond: ",evs[1])
print(len(evs))
frames of first cond: [array([44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60,
61, 62, 63, 64, 65, 66, 67, 68]), array([103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127]), array([161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173,
174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185])]
frames of second cond: [array([15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39]), array([73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89,
90, 91, 92, 93, 94, 95, 96, 97]), array([132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144,
145, 146, 147, 148, 149, 150, 151, 152, 153, 154, 155, 156])]
2
idx = 0 # idx = 0 for 'fear' condition in 'EMOTION task' and idx = 1 for 'neut'
evs[idx]
for i in range(len(evs[idx])):
print(evs[idx][i])
[44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67
68]
[103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120
121 122 123 124 125 126 127]
[161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178
179 180 181 182 183 184 185]
print(data.shape[1])
176
# print(evs[0])
# print(data.shape[1])
idx = 0
frames_cond = evs[idx]
print("before removing",frames_cond)
for i, frame in enumerate(frames_cond):
if frame[-1] > data.shape[1]: #if the time index in frame is larger than the datashape, exclude that frame
print("index where it exceeds data frame: ",i)
frames_cond = np.delete(frames_cond,i, axis=0)
print("after removing the elements",frames_cond)
before removing [array([44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60,
61, 62, 63, 64, 65, 66, 67, 68]), array([103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127]), array([161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173,
174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185])]
index where it exceeds data frame: 2
after removing the elements [[ 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61
62 63 64 65 66 67 68]
[103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120
121 122 123 124 125 126 127]]
print(type(frames_cond[0]))
<class 'numpy.ndarray'>
For the motor task, this evs variable contains a list of 5 arrays corresponding to the 5 conditions.
Now let’s use these evs to compare the average activity during the left foot (‘lf’) and right foot (‘rf’) conditions:
##Back to the core code
# we need a little function that averages all frames from any given condition
def average_frames(data,evs,experiment,cond):
# get idx for the specific condition (ex. fear/neut)
idx = EXPERIMENTS[experiment]['cond'].index(cond)
# get rid of the index that exceed the dataframe
frames_cond = evs[idx]
for i, frame in enumerate(frames_cond):
if frame[-1] > data.shape[1]: #if the time index in frame is larger than the datashape, exclude that frame
frames_cond = np.delete(frames_cond,i, axis=0) #note the frams_cond is still np.array
return np.mean(np.concatenate([np.mean(data[:, frames_cond[i]], axis=1, keepdims=True) for i in range(len(frames_cond))], axis=-1), axis=1)
fear_activity = average_frames(data, evs, my_exp, 'fear')
neut_activity = average_frames(data,evs, my_exp, 'neut')
contrast_emotion = fear_activity-neut_activity
#print(fear_activity)
#fear_activity = average_frames(data, evs, my_exp, 'fear')
#contrast = lf_activity - rf_activity # difference between left and right hand movement
# Plot activity level in each ROI for both conditions
plt.plot(fear_activity,label='fear')
plt.plot(neut_activity,label='neutral')
plt.xlabel('ROI')
plt.ylabel('activity')
plt.legend()
plt.show()
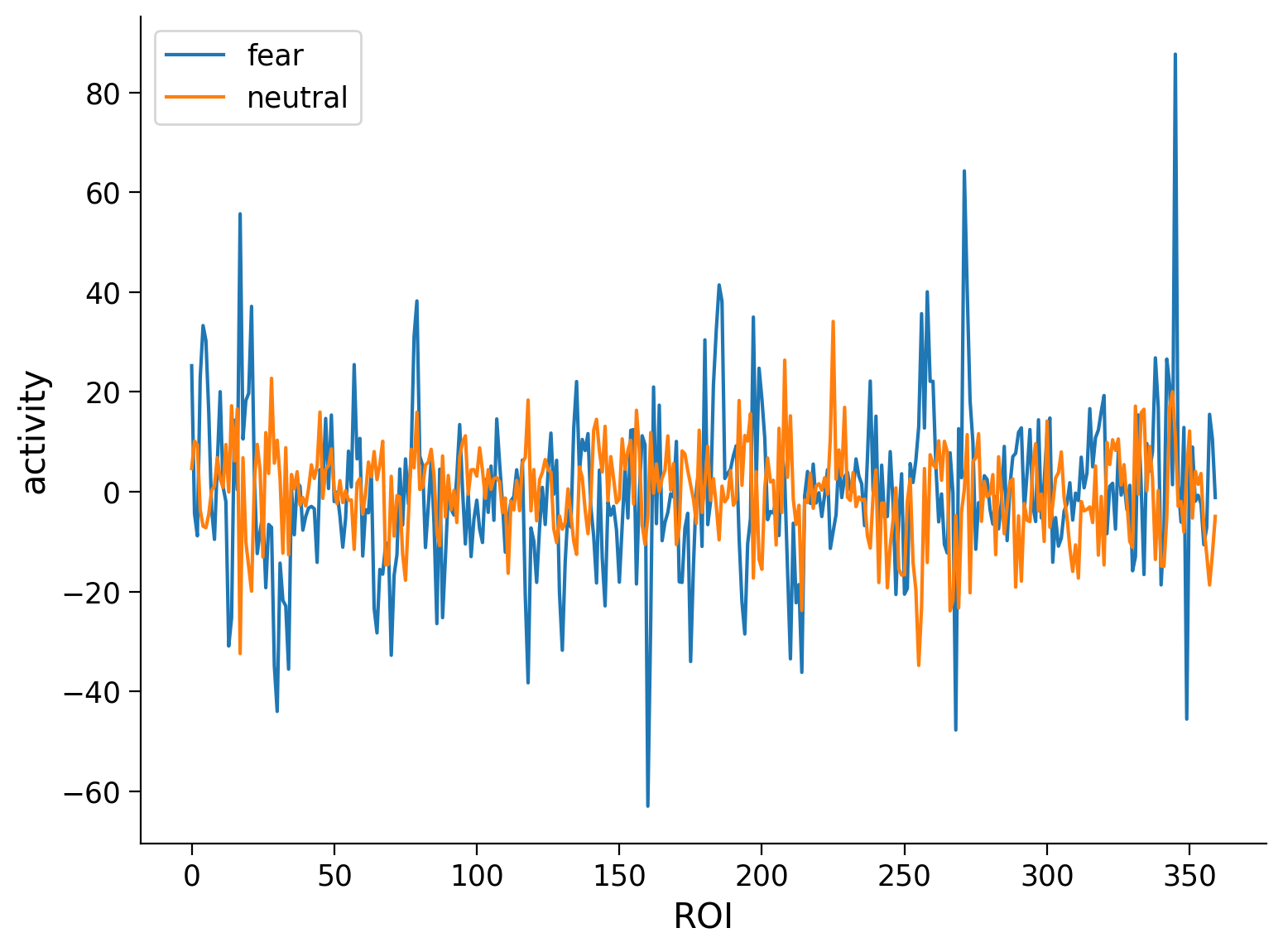
📌Data1: Average BOLD activity for individual for each condition and contrast#
import numpy as np
arr_1D = np.array([1 ,2, 3, 4, 5, 6, 7, 8])
arr_1D = np.delete(arr_1D, np.where(arr_1D == 8))
print(arr_1D)
[1 2 3 4 5 6 7]
print(subjects)
['100307' '100408' '101915' '102816' '103414' '103515' '103818' '105115'
'105216' '106016' '106319' '110411' '111009' '111312' '111514' '111716'
'113215' '113619' '114924' '115320' '117122' '117324' '118730' '118932'
'119833' '120111' '120212' '122317' '123117' '124422' '125525' '126325'
'127933' '128632' '129028' '130013' '130316' '130922' '131924' '133625'
'133827' '133928' '134324' '135932' '136833' '137128' '138231' '138534'
'139637' '140824' '142828' '143325' '148032' '148335' '149337' '149539'
'150524' '151223' '151526' '151627' '153025' '153429' '154431' '156233'
'156637' '157336' '158035' '158540' '159239' '159340' '160123' '161731'
'162329' '162733' '163129' '163432' '167743' '172332' '175439' '176542'
'178950' '182739' '185139' '188347' '189450' '190031' '192439' '192540'
'193239' '194140' '196144' '196750' '197550' '198451' '199150' '199655'
'200614' '201111' '201414' '205119']
print(len(subjects))
100
exc_subjects = np.delete(subjects,np.where(subjects == '133827'))
exc_subjects = np.delete(exc_subjects,np.where(subjects == '192439'))
print(len(exc_subjects))
98
1.1 EMOTION#
my_exp = 'EMOTION'
#initialize array for keeping the data
list_indiv_BOLD_fear = []
list_indiv_BOLD_neut = []
list_indiv_contrast_emotion = []
for i_subj in exc_subjects:
subj_fear = []
subj_neut = []
for i_run in [0, 1]:
data = load_single_timeseries(subject=i_subj, experiment=my_exp,
run=i_run, remove_mean=True)
evs = load_evs(subject=i_subj, experiment=my_exp,run=i_run)
#cond: fear
fear_activity = average_frames(data, evs, my_exp, 'fear')
subj_fear.append(fear_activity)
#cond: neutral
neut_activity = average_frames(data, evs, my_exp, 'neut')
subj_neut.append(neut_activity)
#average of 2 runs and append to list_indiv_BOLD
list_indiv_BOLD_fear.append(np.mean(subj_fear,axis=0))
list_indiv_BOLD_neut.append(np.mean(subj_neut,axis=0))
list_indiv_contrast_emotion.append(np.mean(subj_fear,axis=0) - np.mean(subj_neut,axis=0))
###Emotion Network (contrast only)
#GET Network
my_exp = 'EMOTION'
#initialize array for keeping the data
list_contrast_net_emotion= []
for i_subj in exc_subjects:
subj_fear = []
subj_neut = []
for i_run in [0, 1]:
data = load_single_timeseries(subject=i_subj, experiment=my_exp,
run=i_run, remove_mean=True)
evs = load_evs(subject=i_subj, experiment=my_exp,run=i_run)
#cond: fear
fear_activity = average_frames(data, evs, my_exp, 'fear')
subj_fear.append(fear_activity)
#cond: neutral
neut_activity = average_frames(data, evs, my_exp, 'neut')
subj_neut.append(neut_activity)
#average of 2 runs and append to list_indiv_BOLD
subj_fear_avg = np.mean(subj_fear,axis=0)
subj_neut_avg = np.mean(subj_neut,axis=0)
#Turn 360 regions into 12 networks; do not average over hemisphere so in the end each subj will have 24 features
#Dataframe for emotion
df_emotion = pd.DataFrame({'fear_activity' : subj_fear_avg,
'neut_activity' : subj_neut_avg,
'network' : region_info['network'],
'hemi' : region_info['hemi']})
#fear
list_fear_net = df_emotion.groupby(["network","hemi" ])["fear_activity"].mean().tolist()
#neut
list_neut_net = df_emotion.groupby(["network","hemi" ])["neut_activity"].mean().tolist()
#contrast net
list_contrast_net_emotion.append([list_fear_net[i] - list_neut_net[i] for i in range(len(list_neut_net)) ])
#Check
print(len(list_contrast_net_emotion))
print(len(list_contrast_net_emotion[0]))
98
24
# df_emotion.groupby(["network","hemi" ])["fear_activity"].mean()
network hemi
Auditory Left 10.081486
Right 15.384364
Cingulo-Oper Left 5.887301
Right 2.643987
Default Left 13.471703
Right 10.622781
Dorsal-atten Left 12.426185
Right 5.293913
Frontopariet Left 13.901216
Right 17.991914
Language Left 8.173584
Right 17.617729
Orbito-Affec Left 16.362953
Right 13.828290
Posterior-Mu Left 13.223771
Right 11.458556
Somatomotor Left 5.805786
Right 9.444462
Ventral-Mult Left 26.618241
Right 18.130791
Visual1 Left 9.248596
Right 30.443386
Visual2 Left 17.406913
Right 23.369150
Name: fear_activity, dtype: float64
1.3 GAMBLING task#
my_exp = 'GAMBLING'
#initialize array for keeping the data
list_indiv_BOLD_win = []
list_indiv_BOLD_loss = []
list_indiv_contrast_gambling = []
for i_subj in exc_subjects:
subj_win = []
subj_loss = []
for i_run in [0, 1]:
data = load_single_timeseries(subject=i_subj, experiment=my_exp,
run=i_run, remove_mean=True)
evs = load_evs(subject=i_subj, experiment=my_exp,run=i_run)
#cond: mental
win_activity = average_frames(data, evs, my_exp, 'win')
subj_win.append(win_activity)
#cond: random
loss_activity = average_frames(data, evs, my_exp, 'loss')
subj_loss.append(loss_activity)
#average of 2 runs and append to list_indiv_BOLD
list_indiv_BOLD_win.append(np.mean(subj_win,axis=0))
list_indiv_BOLD_loss.append(np.mean(subj_loss,axis=0))
list_indiv_contrast_gambling.append(np.mean(subj_win,axis=0) - np.mean(subj_loss,axis=0))
###Gambling Network (contrast only)
#GET Network
my_exp = 'GAMBLING'
#initialize array for keeping the data
list_contrast_net_gambling= []
for i_subj in exc_subjects:
subj_win = []
subj_loss = []
for i_run in [0, 1]:
data = load_single_timeseries(subject=i_subj, experiment=my_exp,
run=i_run, remove_mean=True)
evs = load_evs(subject=i_subj, experiment=my_exp,run=i_run)
#cond: mental
win_activity = average_frames(data, evs, my_exp, 'win')
subj_win.append(win_activity)
#cond: random
loss_activity = average_frames(data, evs, my_exp, 'loss')
subj_loss.append(loss_activity)
#average of 2 runs and append to list_indiv_BOLD
subj_win_avg = np.mean(subj_win,axis=0)
subj_loss_avg = np.mean(subj_loss,axis=0)
#Turn 360 regions into 12 networks; do not average over hemisphere so in the end each subj will have 24 features
#Dataframe for emotion
df_gambling = pd.DataFrame({'win_activity' : subj_win_avg,
'loss_activity' : subj_loss_avg,
'network' : region_info['network'],
'hemi' : region_info['hemi']})
#win
list_win_net = df_gambling.groupby(["network","hemi" ])["win_activity"].mean().tolist()
#loss
list_loss_net = df_gambling.groupby(["network","hemi" ])["loss_activity"].mean().tolist()
#contrast net
list_contrast_net_gambling.append([list_win_net[i] - list_loss_net[i] for i in range(len(list_win_net)) ])
#Check
print(len(list_contrast_net_gambling))
print(len(list_contrast_net_gambling[0]))
98
24
Writing csv files#
#network columns
network_columns = ['Auditory_L','Auditory_R','Cingulo-Oper_L','Cingulo-Oper_R','Default_L','Default_R', 'Dorsal-atten_L','Dorsal-atten_R', 'Frontopariet_L', 'Frontopariet_R', 'Language_L', 'Language_R' , 'Orbito-Affec_L', 'Orbito-Affec_R', 'Posterior-Mu_L', 'Posterior-Mu_R', 'Somatomotor_L', 'Somatomotor_R', 'Ventral-Mult_L', 'Ventral-Mult_R', 'Visual1_L', 'Visual1_R', 'Visual2_L', 'Visual2_L']
#get col name
# let's assume these lists have been populated with the relevant sections
list_task = ['Emotion','Social','Gambling']
for i in range(len(network_columns)):
col_emotion = [list_task[0]+'_'+area for area in network_columns]
for i in range(len(network_columns)):
col_social = [list_task[1]+'_'+area for area in network_columns]
for i in range(len(network_columns)):
col_gambling = [list_task[2]+'_'+area for area in network_columns]
#check
print(col_emotion)
print(col_social)
print(col_gambling)
list_columns_net = ['subject_id'] + col_emotion + col_social + col_gambling
print(len(list_columns_net) == 24*3+1)
['Emotion_Auditory_L', 'Emotion_Auditory_R', 'Emotion_Cingulo-Oper_L', 'Emotion_Cingulo-Oper_R', 'Emotion_Default_L', 'Emotion_Default_R', 'Emotion_Dorsal-atten_L', 'Emotion_Dorsal-atten_R', 'Emotion_Frontopariet_L', 'Emotion_Frontopariet_R', 'Emotion_Language_L', 'Emotion_Language_R', 'Emotion_Orbito-Affec_L', 'Emotion_Orbito-Affec_R', 'Emotion_Posterior-Mu_L', 'Emotion_Posterior-Mu_R', 'Emotion_Somatomotor_L', 'Emotion_Somatomotor_R', 'Emotion_Ventral-Mult_L', 'Emotion_Ventral-Mult_R', 'Emotion_Visual1_L', 'Emotion_Visual1_R', 'Emotion_Visual2_L', 'Emotion_Visual2_L']
['Social_Auditory_L', 'Social_Auditory_R', 'Social_Cingulo-Oper_L', 'Social_Cingulo-Oper_R', 'Social_Default_L', 'Social_Default_R', 'Social_Dorsal-atten_L', 'Social_Dorsal-atten_R', 'Social_Frontopariet_L', 'Social_Frontopariet_R', 'Social_Language_L', 'Social_Language_R', 'Social_Orbito-Affec_L', 'Social_Orbito-Affec_R', 'Social_Posterior-Mu_L', 'Social_Posterior-Mu_R', 'Social_Somatomotor_L', 'Social_Somatomotor_R', 'Social_Ventral-Mult_L', 'Social_Ventral-Mult_R', 'Social_Visual1_L', 'Social_Visual1_R', 'Social_Visual2_L', 'Social_Visual2_L']
['Gambling_Auditory_L', 'Gambling_Auditory_R', 'Gambling_Cingulo-Oper_L', 'Gambling_Cingulo-Oper_R', 'Gambling_Default_L', 'Gambling_Default_R', 'Gambling_Dorsal-atten_L', 'Gambling_Dorsal-atten_R', 'Gambling_Frontopariet_L', 'Gambling_Frontopariet_R', 'Gambling_Language_L', 'Gambling_Language_R', 'Gambling_Orbito-Affec_L', 'Gambling_Orbito-Affec_R', 'Gambling_Posterior-Mu_L', 'Gambling_Posterior-Mu_R', 'Gambling_Somatomotor_L', 'Gambling_Somatomotor_R', 'Gambling_Ventral-Mult_L', 'Gambling_Ventral-Mult_R', 'Gambling_Visual1_L', 'Gambling_Visual1_R', 'Gambling_Visual2_L', 'Gambling_Visual2_L']
True
import csv
import pandas as pd
list_all_data = []
for row_idx in range(len(list_contrast_net_emotion)):
row = []
row.append(exc_subjects[row_idx])
row.extend(list_contrast_net_emotion[row_idx])
row.extend(list_contrast_net_social[row_idx])
row.extend(list_contrast_net_gambling[row_idx])
list_all_data.append(row)
df_BOLD = pd.DataFrame(list_all_data,columns=list_columns_net)
print(df_BOLD)
df_BOLD.to_csv('BOLD_contrast_network_98.csv', index=False)
subject_id Emotion_Auditory_L Emotion_Auditory_R Emotion_Cingulo-Oper_L \
0 100307 -2.804921 -2.433804 -0.777985
1 100408 3.516186 7.529671 -9.521039
2 101915 9.980518 5.266869 9.546204
3 102816 20.833271 -16.487274 1.886792
4 103414 9.885700 5.633930 9.734162
.. ... ... ... ...
93 199655 -3.796242 -3.248363 -12.931731
94 200614 -2.340933 -0.460370 -6.904875
95 201111 -0.313696 -3.006372 7.014663
96 201414 9.047621 3.032789 -4.672088
97 205119 18.881525 12.401503 7.964594
Emotion_Cingulo-Oper_R Emotion_Default_L Emotion_Default_R \
0 -0.353353 10.053771 5.986613
1 -5.284645 -8.731886 7.884268
2 11.319655 8.908589 20.103598
3 -0.946059 10.643481 3.247931
4 9.824339 13.819462 21.356125
.. ... ... ...
93 -7.968932 -1.838899 -2.248285
94 -13.213405 -0.929012 -7.979797
95 -1.106524 7.094315 1.009450
96 -2.014852 0.149621 22.042231
97 -0.326323 24.005220 15.183962
Emotion_Dorsal-atten_L Emotion_Dorsal-atten_R Emotion_Frontopariet_L \
0 11.420777 8.673832 4.628537
1 11.791724 1.867515 -8.937923
2 0.907064 12.911264 2.576694
3 13.517639 13.682730 -7.682030
4 21.945681 7.596248 13.637148
.. ... ... ...
93 -0.109514 1.404556 -8.534893
94 11.367749 18.012441 -1.613193
95 9.850395 6.223135 2.587723
96 1.823159 22.238250 -1.161439
97 16.718064 9.807230 15.608610
... Gambling_Posterior-Mu_L Gambling_Posterior-Mu_R \
0 ... -7.973004 2.530932
1 ... -24.315753 -18.506888
2 ... -1.944254 -0.950986
3 ... -3.673315 -2.800902
4 ... 9.498112 7.589702
.. ... ... ...
93 ... -4.010796 1.193511
94 ... 11.388163 11.611773
95 ... -1.079248 10.458447
96 ... 4.350870 2.288127
97 ... -15.698724 -16.134373
Gambling_Somatomotor_L Gambling_Somatomotor_R Gambling_Ventral-Mult_L \
0 -3.179724 -1.810594 -7.124124
1 -6.057032 4.568156 -48.646206
2 0.150241 3.160913 -8.325856
3 12.236763 6.242775 -9.369718
4 4.858379 14.831399 21.459354
.. ... ... ...
93 0.801685 -0.544744 -0.791156
94 -12.670486 4.437090 1.793672
95 -10.125810 -8.503565 6.909634
96 -0.454990 7.512785 10.580950
97 -19.458602 -16.554394 -16.920817
Gambling_Ventral-Mult_R Gambling_Visual1_L Gambling_Visual1_R \
0 1.524933 -3.339643 -3.025932
1 -11.067792 -16.796800 -5.983401
2 -9.110250 -7.625908 -1.689586
3 -12.720637 8.636415 20.621203
4 -7.949689 18.820264 10.545901
.. ... ... ...
93 22.397713 -2.309791 9.854299
94 -0.470018 -8.310730 9.031029
95 26.248419 15.927485 13.441762
96 7.709618 0.995246 0.200687
97 -41.737430 14.800047 19.709679
Gambling_Visual2_L Gambling_Visual2_L
0 8.826484 7.199637
1 -8.297101 6.657371
2 -1.384789 1.175621
3 9.931208 8.350895
4 12.616854 12.111169
.. ... ...
93 -1.874123 0.729667
94 -6.830466 -0.281131
95 6.393254 4.603233
96 3.556047 14.316392
97 -5.829851 -10.290764
[98 rows x 73 columns]
Columns for 360 areas#
#get col name
# let's assume these lists have been populated with the relevant sections
list_columns = []
list_columns.append("subject_id")
for i in range(360):
list_columns.append(f"BOLD_fear_{i+1}")
for i in range(360):
list_columns.append(f"BOLD_neut_{i+1}")
for i in range(360):
list_columns.append(f"contrast_emotion_{i+1}")
for i in range(360):
list_columns.append(f"BOLD_mental_{i+1}")
for i in range(360):
list_columns.append(f"BOLD_random_{i+1}")
for i in range(360):
list_columns.append(f"contrast_social_{i+1}")
for i in range(360):
list_columns.append(f"BOLD_win_{i+1}")
for i in range(360):
list_columns.append(f"BOLD_loss_{i+1}")
for i in range(360):
list_columns.append(f"contrast_gambling_{i+1}")
print(len(list_columns) == 360*9 +1)
True
import csv
import pandas as pd
list_all_data = []
for row_idx in range(len(list_indiv_BOLD_fear)):
row = []
row.append(exc_subjects[row_idx])
row.extend(list_indiv_BOLD_fear[row_idx])
row.extend(list_indiv_BOLD_neut[row_idx])
row.extend(list_indiv_contrast_emotion[row_idx])
row.extend(list_indiv_BOLD_mental[row_idx])
row.extend(list_indiv_BOLD_random[row_idx])
row.extend(list_indiv_contrast_social[row_idx])
row.extend(list_indiv_BOLD_win[row_idx])
row.extend(list_indiv_BOLD_loss[row_idx])
row.extend(list_indiv_contrast_gambling[row_idx])
list_all_data.append(row)
df_BOLD = pd.DataFrame(list_all_data,columns=list_columns)
df_BOLD.to_csv('BOLD_98_individual.csv', index=False)
from google.colab import drive
drive.mount('/content/drive')
SOCIAL Task#
#SOCIAL task average frame calculate
my_exp = 'SOCIAL'
my_subj = subjects[0]
my_run = 1
#use load_single_timeseries
data = load_single_timeseries(subject=my_subj,
experiment=my_exp,
run=my_run,
remove_mean=True)
#use load_evs
evs = load_evs(subject=my_subj, experiment=my_exp, run=my_run)
#use average frame
rand_activity = average_frames(data, evs, my_exp, 'rnd')
mental_activity = average_frames(data,evs, my_exp, 'mental')
# SOCIAL: Plot activity level in each ROI for both conditions
plt.plot(rand_activity,label='random')
plt.plot(mental_activity,label='mental')
plt.xlabel('ROI')
plt.ylabel('activity')
plt.legend()
plt.show()
GAMBLING task#
#GAMBLING task average frame calculate
my_exp = 'GAMBLING'
my_subj = subjects[0]
my_run = 1
#use load_single_timeseries
data = load_single_timeseries(subject=my_subj,
experiment=my_exp,
run=my_run,
remove_mean=True)
#use load_evs
evs = load_evs(subject=my_subj, experiment=my_exp, run=my_run)
#use average frame
win_activity = average_frames(data, evs, my_exp, 'win')
loss_activity = average_frames(data,evs, my_exp, 'loss')
# GAMBLING: Plot activity level in each ROI for both conditions
plt.plot(win_activity,label='win')
plt.plot(loss_activity,label='loss')
plt.xlabel('ROI')
plt.ylabel('activity')
plt.legend()
plt.show()
Try plotting the “average frame” data of ALL subjects#
# try plotting Brain activity for each tasks
# Gambling
my_exp = 'GAMBLING'
subj_no = 1
for cond in EXPERIMENTS[my_exp]['cond']:
list_activity = [] # initializing the list_ activity
for subj_i in range(len(subjects)):
my_subj = subjects[subj_i]
my_run = 1
data = load_single_timeseries(subject=my_subj,
experiment=my_exp,
run=my_run,
remove_mean=True)
evs = load_evs(subject=my_subj, experiment=my_exp, run=my_run)
gambling_activity = average_frames(data,evs,my_exp,cond)
# Plot activity level in each ROI for both conditions
list_activity.append(gambling_activity)
subj_no += 1
plt.plot(list_activity)
plt.xlabel('ROI')
plt.ylabel('activity')
plt.legend()
plt.title(cond)
plt.show()
# try plotting Brain activity for each tasks
#
my_exp = 'SOCIAL'
subj_no = 1
for cond in EXPERIMENTS[my_exp]['cond']:
list_activity = [] # initializing the list_ activity
for subj_i in range(len(subjects)):
my_subj = subjects[subj_i]
my_run = 1
data = load_single_timeseries(subject=my_subj,
experiment=my_exp,
run=my_run,
remove_mean=True)
evs = load_evs(subject=my_subj, experiment=my_exp, run=my_run)
gambling_activity = average_frames(data,evs,my_exp,cond)
# Plot activity level in each ROI for both conditions
list_activity.append(gambling_activity)
subj_no += 1
plt.plot(list_activity)
plt.xlabel('ROI')
plt.ylabel('activity')
plt.legend()
plt.title(cond)
plt.show()
# plot according to the network
avg_activity = np.mean(list_activity)
print('average',avg_activity)
# plt.plot(avg_activity)
# plt.xlabel('ROI')
# plt.ylabel('activity')
# plt.legend()
# plt.title('average')
# plt.show()
# df = pd.DataFrame({'neutral_activity' : avg_activity,
# 'network' : region_info['network'],
# 'hemi' : region_info['hemi']})
# sns.barplot(y='network', x='neutral_activity', data=df, hue='hemi')
# plt.show()
#try running through all the data and subjects to check if all the data can be accessed
subj_no = 1
for subj_i in range(len(subjects)):
for experiment, conds in EXPERIMENTS.items():
for cond in conds['cond']:
my_exp = experiment
my_subj = subjects[subj_i]
my_run = 1
data = load_single_timeseries(subject=my_subj,
experiment=my_exp,
run=my_run,
remove_mean=True)
evs = load_evs(subject=my_subj, experiment=my_exp, run=my_run)
try:
average_frames(data,evs,my_exp,cond)
except:
print('error: ', str(subj_i), experiment, cond)
subj_no += 1
Contrast and network#
Now let’s plot these activity vectors. We will also make use of the ROI names to find out which brain areas show highest activity in these conditions. But since there are so many areas, we will group them by network.
A powerful tool for organising and plotting this data is the combination of pandas and seaborn. Below is an example where we use pandas to create a table for the activity data and we use seaborn oto visualise it.
#EMOTION task
df = pd.DataFrame({'fear_activity' : fear_activity,
'neut_activity' : neut_activity,
'network' : region_info['network'],
'hemi' : region_info['hemi']})
fig, (ax1, ax2) = plt.subplots(1, 2)
sns.barplot(y='network', x='fear_activity', data=df, hue='hemi',ax=ax1)
sns.barplot(y='network', x='neut_activity', data=df, hue='hemi',ax=ax2)
plt.show()
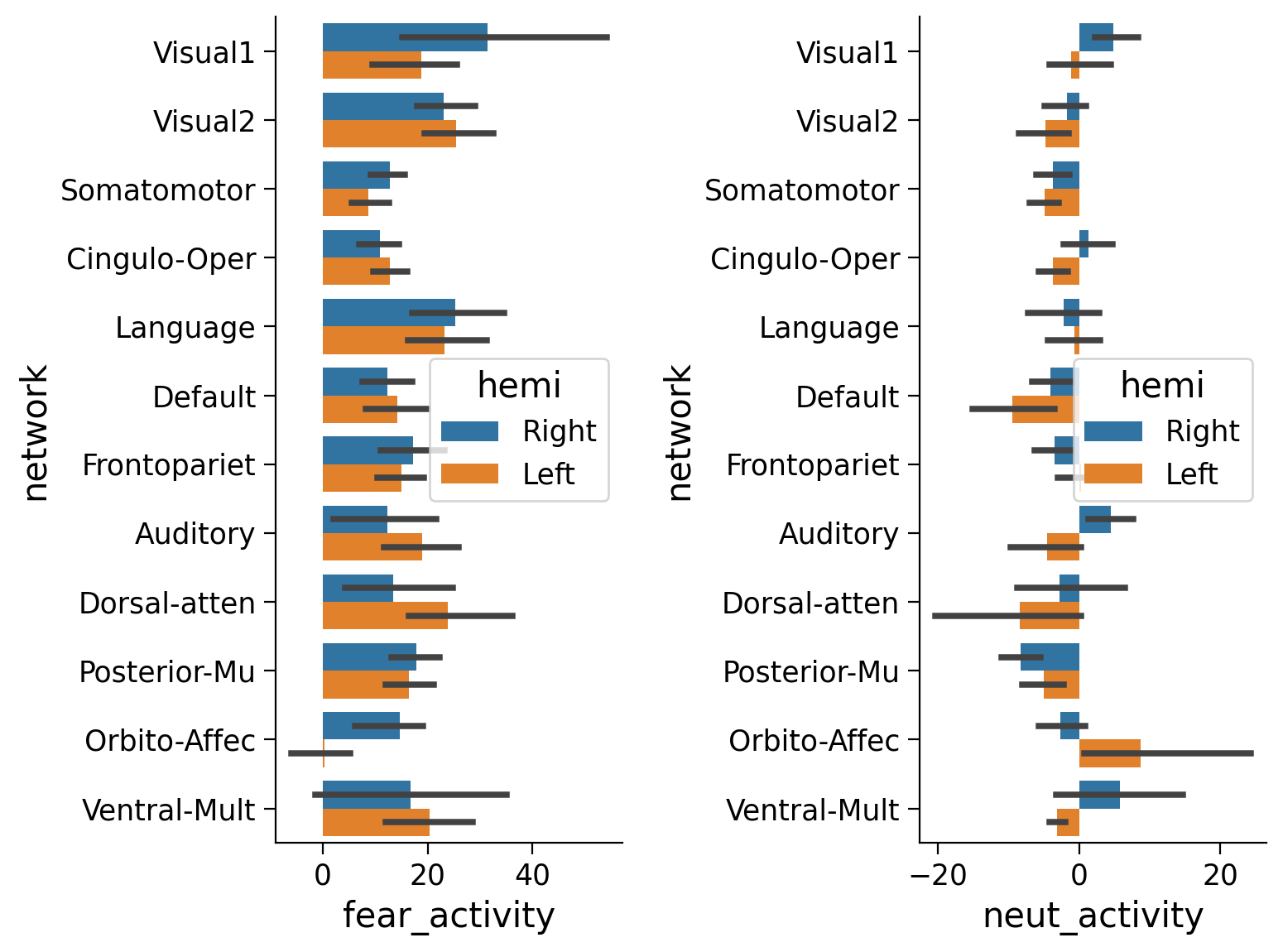
#SOCIAL task
df = pd.DataFrame({'mental_activity' : mental_activity,
'rand_activity' : rand_activity,
'network' : region_info['network'],
'hemi' : region_info['hemi']})
print(df)
fig, (ax1, ax2) = plt.subplots(1, 2)
sns.barplot(y='network', x='mental_activity', data=df, hue='hemi',ax=ax1) #sns barplot prob. show only 10 first regions
sns.barplot(y='network', x='rand_activity', data=df, hue='hemi',ax=ax2)
plt.show()
You should be able to notice that for the somatosensory network, brain activity in the right hemi is higher for the left foot movement and vice versa for the left hemi and right foot. But this may be subtle at the single subject/session level (these are quick 3-4min scans).
Let us boost thee stats by averaging across all subjects and runs.
Average across all subjects and runs#
#EMOTION task
#EMOTION
my_exp = 'EMOTION' #'SOCIAL' , 'GAMBLING'
group_contrast_emotion = 0
for s in subjects:
for r in [0, 1]:
data = load_single_timeseries(subject=s, experiment=my_exp,
run=r, remove_mean=True)
evs = load_evs(subject=s, experiment=my_exp,run=r)
fear_activity = average_frames(data, evs, my_exp, 'fear')
neut_activity = average_frames(data, evs, my_exp, 'neut')
contrast = fear_activity - neut_activity
group_contrast_emotion += contrast
group_contrast_emotion /= (len(subjects)*2) # remember: 2 sessions per subject
df = pd.DataFrame({'contrast' : group_contrast_emotion,
'network' : region_info['network'],
'hemi' : region_info['hemi']
})
# we will plot the fear foot minus neutral foot contrast so we only need one plot
plt.figure()
sns.barplot(y='network', x='contrast', data=df, hue='hemi')
plt.title('group contrast of fear vs neutral')
plt.show()
Visualising the results on a brain#
Finally, we will visualise these resuts on the cortical surface of an average brain.
# @title NMA provides an atlas. Run this cell to download it
import os, requests
# NMA provides an atlas
fname = f"{HCP_DIR}/atlas.npz"
url = "https://osf.io/j5kuc/download"
if not os.path.isfile(fname):
try:
r = requests.get(url)
except requests.ConnectionError:
print("!!! Failed to download data !!!")
else:
if r.status_code != requests.codes.ok:
print("!!! Failed to download data !!!")
else:
with open(fname, "wb") as fid:
fid.write(r.content)
with np.load(fname) as dobj:
atlas = dict(**dobj)
# This uses the nilearn package
from nilearn import plotting, datasets
# Try both hemispheres (L->R and left->right)
fsaverage = datasets.fetch_surf_fsaverage()
surf_contrast = group_contrast_emotion[atlas["labels_L"]]
plotting.view_surf(fsaverage['infl_left'],
surf_contrast,
vmax=20)
GAMBLING#
#GAMBLING
my_exp = 'GAMBLING'
#calculate group contrast GAMBLING
group_contrast_gambling = 0
for s in subjects:
for r in [0, 1]:
data = load_single_timeseries(subject=s, experiment=my_exp,
run=r, remove_mean=True)
evs = load_evs(subject=s, experiment=my_exp,run=r)
win_activity = average_frames(data, evs, my_exp, 'win')
loss_activity = average_frames(data, evs, my_exp, 'loss')
contrast = win_activity - loss_activity
group_contrast_gambling += contrast
group_contrast_gambling /= (len(subjects)*2) # remember: 2 sessions per subject
#Plot contrast bar graph GAMBLING
df = pd.DataFrame({'contrast' : group_contrast_gambling,
'network' : region_info['network'],
'hemi' : region_info['hemi']
})
# we will plot the fear foot minus neutral foot contrast so we only need one plot
plt.figure()
sns.barplot(y='network', x='contrast', data=df, hue='hemi')
plt.title('group contrast of win vs loss')
plt.show()
#PLot the on atlas SOCIAL
# Try both hemispheres (L->R and left->right)
fsaverage = datasets.fetch_surf_fsaverage()
surf_contrast = group_contrast_gambling[atlas["labels_L"]]
plotting.view_surf(fsaverage['infl_left'],
surf_contrast,
vmax=20)
Archived Codes#
def average_frames_modi(data, evs, experiment, cond):
idx = EXPERIMENTS[experiment]['cond'].index(cond) #the index of condition ex. 'fear' = 0, 'neut = 1' this depends on the position of that condition in dict EXPERIMENT
list_of_mean = []
for i in range(len(evs[idx])-1):
print("len(data[:, evs[idx][i]])",len(data[:, evs[idx][i]]))
print("evs[idx][i]",evs[idx][i])
print("data[:, evs[idx][i]]", data[:, evs[idx][i]])
list_of_mean.append(np.mean(data[:, evs[idx][i]], axis=1, keepdims=True))
print("len(list_of_mean)",len(list_of_mean)) # ==> the list of mean is a list(2) of lists (360); this is the same as the result of list comprehension below
print("len(list_of_mean[0])",len(list_of_mean[0]))
#print("list_of_mean",list_of_mean)
np.concatenate(list_of_mean,axis=-1)
print("len(np.concatenate(list_of_mean,axis=-1))",len(np.concatenate(list_of_mean,axis=-1)))
print("np.concatenate(list_of_mean,axis=-1)",np.concatenate(list_of_mean,axis=-1))
print("len(np.mean(np.concatenate(list_of_mean), axis=1))",len(np.mean(np.concatenate(list_of_mean), axis=1)))
print("np.mean(np.concatenate(list_of_mean), axis=1)",np.mean(np.concatenate(list_of_mean), axis=1))
return np.mean(np.concatenate(list_of_mean), axis=1)


1.2 SOCIAL task#
###Social Network (contrast only)